|
2.0.0b10
catchment modelling framework
|
|
2.0.0b10
catchment modelling framework
|
Every flux and every state can be accessed during runtime. Thus you can save anything you like to the output during the run time loop. In the following we will have a look at some techniques how to see what is going on in a model. As an example model we use the very simple 2 storage model with boundary condition from CmfTutBoundary.
To show our results, we are using Python's print statement / function. However, "print" has changed between python 2 and 3. To run our examples python 2 users should always start their programs with the following line:
This import the behaviour of the python3 print function as well as the improved handling of integer division into python 2. Using this line in python3 is fine and changes nothing.
We have already asked for states in the example model. Every water storage has the property volume to return the stored water volume in m³. More general, state returns the value stored in a state variable, which is usually volume for a water storage and the tracer mass for a solute storage. Water storages with a defined relation between head and volume (such storages will be covered later, eg. SoilLayer and OpenWaterStorage), can also give back their potential.
The properties can also be used to change the states of the system, but this is only useful for initial conditions. Changing the state during runtime will of course destroy the water balance. Some of the solvers will even ignore the changes if they are not reset. In our simple example we have used this already for the initial condition:
Getting fluxes is a little bit more complicated than getting states: Some fluxes depend only on the state of the connected water storages, like LinearStorageConnection, some on time, like the NeumannBoundary and some on both. Since the storages / nodes do not know, what time we have (this is a property of the solver) you need to give the time into the query.
Fluxes between two nodes source and target at time t can be queried with the flux_to method:
For our toy model you can query all fluxes the following way:
You get the result of the actual flux between source and target in \(m^3/day\). If water is flowing from source to target, the result is positive else negative. If source and target are not connected you will get 0.0, and no error will occur. Since you get the actual flux, summing up the fluxes does not necessarily reflect the true accumulated flux, since in non-linear models, the integrated flux over a timestep does not equal the sum. CMF is very mass conservative, since the water volume is the integrated state variable, but by summing up you might get the impression of numerical errors in the integration.
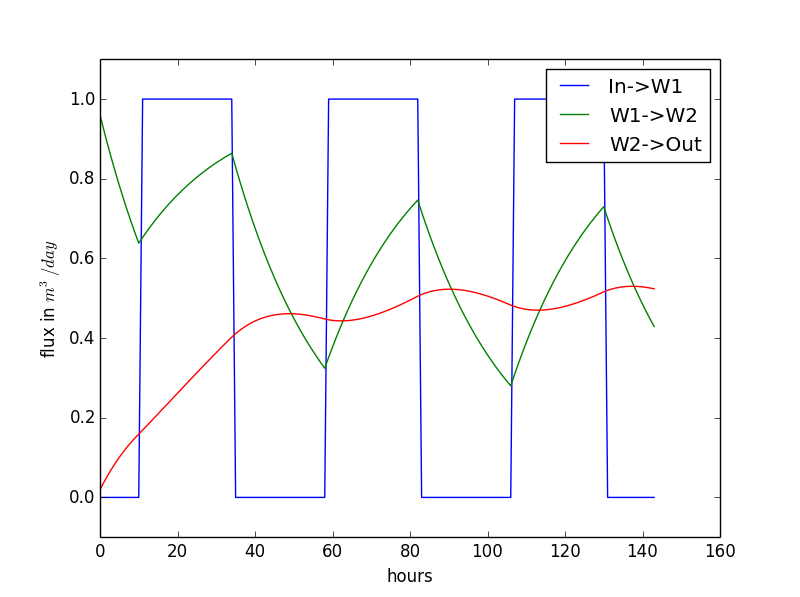
Now we are going to save the fluxes instead of the volume in a list in our first model with boundary conditions and plot the result:
The result of this code is on the right.
You can get all fluxes of one node with target.fluxes(t). You get a list of tuples, each tuple composed of the flux and the target node. Note: positive fluxes here mean fluxes into the target.
Example:
Returns:
If you want to know the current balance (sum) of the fluxes you can of course do:
But you can also let cmf do the sum:
or even shorter:
This is especially useful for boundary conditions, however this does also only show the actual waterbalance and not the average waterbalance over a given timestep. Hence accumulating the waterbalance over time leads to numerical errors.
If you need the exact waterbalance, you can create another waterstorage and connect the boundary condition with an WaterbalanceFlux
Sometimes, you might not be sure what storages are exactly connected to each other, or the following tools are just simpler to use, then inspecting your setup code.
Getting a list of all the connections of one node is done by the connections property of a node (storage, boundary).
Example:
prints:
kinematic wave({W1}<->{W2})
Neumann boundary flux({Boundary at W1}<->{W1})
connected_nodes is a list of the nodes that are connected to another.
prints:
{W2}
{Boundary at W1}